|
|
||
| Menu | OPAL / LynnTsai | |
|
OPAL Home About People International Collaborators
Opportunities
Events
Related Links
|
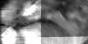
Segmentation of the Soft Palate from MR ImagesBy Lynn Tsai University of British Columbia Prepared for EECE466 Course Project ABSTRACTSegmentation of soft tissue anatomical objects from MR images is a difficult task. The complication comes from the low level of contract at the edges of different anatomical parts in the image and the fact that the soft tissue is prone to higher levels of deformation. In this report a few methods of segmentation methods are discussed in terms of optimizing 3D segmentation of the soft palatal region. These methods include the Region Growing method and the Level Set Method. The usage of real time application Livewire is also included in this report. A few different approaches to using a combination of Livewire and ITK's Geodesic Active Contour Segmentation is compared and discussed. Final results are presented in 2D sample slices and 3D contour maps for the easy visualization of the segmented volume. 1. INTRODUCTION1.1 Background Obstructive Sleep Apnea is a chronic disease that causes degradation of sleep quality, insomnia, and in some cases, death during sleep. The disease is caused by the collapsing of the soft tissue surrounding the airway during sleep when the muscles are relaxed. Because the symptoms of the disease—such as snoring and neurological arousal that may not completely awaken the patient—are often unnoticed by the patient, most people do not realize how common the disease is. It is estimated that at least 10% of all Americans have some form of obstructive sleep apnea. Treatment for obstructive sleep apnea includes the C-CAP or the Bi-CAP(Continuous /Bi- Constant Airway Pressure), which is a machine worn around the patient’s head to keep the airway open using continuous air flow. For patients with more severe forms of the disease, surgery can be done to either intentionally scar the soft tissue to form hard scar tissue to give the airway more rigid structural support or remove excess tissue surrounding the airway. Usually, parts of the pharynx, uvulo, or palate are removed. Another type of surgery available is the mandibular myotomy, which cuts then reattaches a portion of the jaw to pull the tongue forward so that it does not protrude into the airway. 1.2 Motive for Research Current treatments for the disease are often ineffective because there are no guidelines as to which form of treatment suits the individual. The rate that symptoms reoccur after surgery is high, because often the incorrect type if surgery is prescribed. Each individual’s vocal tract responds differently to air flow and the shape of the tract is deformed differently when the tract is under pressure in the sleeping position. In other words, if surgery does not remove the obstructive tissue but reduces the structural integrity of the airway, then it does more harm than good. Therefore, understanding how each anatomical structure in the airway deforms the airway can provide valuable information to the physician in choosing which surgical operation suits the patient. By reconstructing a 3D model of the soft palate-- a soft tissue structure that is associated with obstructing the airway may provide some insight into solving the problem. 2. THEORY2.1 Region Growing Segmentation Region growing segmentation algorithm uses a seed region and calculates the neighboring pixels to determine if they are a part of the seed region. Therefore, the seed region chosen must be contained in the region of interest to allow the “growing” of the seed outwards. The Confidence Connected segmentation algorithm is an example of region growing segmentation, which uses the following equation: I(X) ∈ [m− fs,m+ fs] *[1] where m and s are the mean and standard deviation of the region intensities, f is a factor defined by the user, I( ) is the image and X is the position of the particular neighbor pixel being considered for inclusion in the region. 2.2 Level Set Segmentation Another method of segmentation used was the level set segmentation algorithm, which starts from a given contour, and expands or contracts the contour by finding a zero level set near by. The level set equation solved is the following partial differential equation: I(X) ∈ [m− fs,m+ fs] *[1] where m and s are the mean and standard deviation of the region intensities, f is a factor defined by the user, I( ) is the image and X is the position of the particular neighbor pixel being considered for inclusion in the region. Geodesic Active Contour Segmentation is an example of level set segmentation method, developed by Leventon et al in *[2]. It utilizes the more common Fast March filter, which is often used to extract objects of known shapes from images. 2.3 Livewire Livewire is a real time user interface written in matlab that enables semi-automatically segmentation a number of 2D contours *[3] . The user is allowed to guide the path of segmentation by gliding the courser over the edge of the region of interest, while Livewire calculates the location of the contours in real time. 3. METHOD3.1 Preprocessing The original data set must be preprocessed before segmentation can be applied. First, a 3D volume data must be reconstructed from the raw data slice set. Then, smoothing and filtering must be applied to the volume to create a continuous 3D image of the original data set. Different methods of preprocessing affects segmentation results significantly, as different segmentation methods require different preprocessed input data volumes and guidance volume images for optimal results. 3.1.1 Reconstruction of Data volume from slices The original MR data set was given as 54 slices of 256x256 pixel images. The slices were stacked along the z-direction and the edges truncated to build a 54 216 pixel x 216 pixel 3D image. Interpolation was used to fill 3 more data points in between every 2 data point in the z direction, giving a final 216x216x216 voxel image. After re-slicing the data volume in an orthogonal direction, it was discovered that the slices were taken in 3 different direction: 1/3 in the coronal direction, 1/3 in the axial direction, and 1/3 ~45 degrees from the coronal plane. The purpose for taking MR slices in this unconventional fashion was to track the shape of the airway, which bends at almost 90 degrees just below the upper airway. 3.1.2 Registration of volumes of different orientations The orientation of the slices makes reconstruction more difficult. Assuming that the 3 sets of data were taken roughly at the same time and that the subject did not move during this time, registration was performed based on mutual information. A rigid body transformation was performed on each volume after correlating regions of overlap in the images. The scaling and rotational factors were determined in a 2D fashion: a sagittal slice was taken from the centre of the each reconstructed volume, and correlation of the 3 sagittal slices was performed with the aid of matlab's “cpselect” toolbox. Using “cpselect”, regions of mutual information was manually selected and inputted into the algorithm in the form of 4 control points. The transformation factors calculated from the single sample slice was then applied to all slices in the volumes. 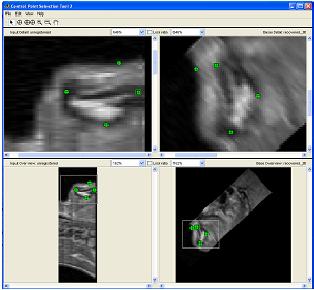 Figure 1: Selecting control points for correlation using mutual information in Matlab’s cpselect Matitk's “RegisterThinPlateSpline” algorithm was also explored. However, this method did not preserve geometries of the original data as well as the crude registration method I had developed, because using matlab's “cpselect” function, the user can graphically select seed points for correlating 2 images, where as in Matitk, a more complex algorithm designed for higher degrees of deformation was used. 3.1.2 Smoothing An output image was created from the overlay of the three registered 3D images. The intensity of overlapped regions was averaged. Then the output was smoothed using Matitk's CurvatureAnsioFilter (FCA) filter to smooth out discontinuities from the registration process. The filter is implemented based on the Perona and Malik's anisotropic diffusion filter. By applying this filter, some of the speckles from the original data slices was also removed. 3.2 Segmentation Segmentation of the soft palate appeared to be very difficult at first. The edges around the palate were hard to recognize even by eye and the shape of the region is irregular. A fully automated method was practically impossible to implement. 2D experiments were first performed on sample slices where contract is high and the features of the palatal region is visible. But results were not satisfactory. Then, with the aid of a semi-automated tool-- Livewire-- better 2D and 3D segmentation results were obtained. 3.2.1 2D Experiments Several segmentation methods were used. First, 2D segmentation was experimented on sample slices. The canny edge detector could successfully segment the airway in the sample slices, since the airway is distinctively low in intensity and surrounded by high intensity regions in the images. Seed points that indicate the approximate location of the region of interest was needed for edge detection. However, the soft palate region could not easily be segmented because the intensity drop off around the edges is not high enough. 3.2.2 3D extension Simply applying a Gaussian filter then segmenting the soft palate region has the same problem: the edges of the soft plate is hard to distinguish even by the human eye. 3.2.2.1 Confidence Connected Segmentation Using the Confidence Connected Segmentation method implemented in Matitk did not give satisfactory results neither. The region growing segmentation methods are not ideal for the soft palate because the algorithm detects intensities based on a threshold value. Again, since the edges are not clearly defined and the intensity inside the region is not homogeneous, results are often inconsistent. Also, seed regions that optimize the result are hard to determine. The results obtained shows the contour of the airway. This proves that the Connected Confidence method can be used to segment regions of homogeneous intensity, as does the airway. 3.2.2.2 Geodesic Active Contour Level Set Segmentation The ITK user manual requires using a gradient map of the feature image as the active contour. Initial experiments with the gradient map was done by using the a Gaussian Magnitude map produced by the Gradient Anisotropic Diffusion filter in Matitk. No results could be obtained. For accurate automated segmentation, a human-in the-loop method is optimal for regions of such complexity. The 2D Livewire was used to determine the active contour supplied to Matitk's Geodesic Active Contour Level Set Segmentation algorithm. 4. RESULTSThree orthogonal slices were chosen on which 2D live wire's semi-automated segmentation was applied. The slices were chosen this way: the same sagittal slice used for registering the volume sets was used first. In the Livewire UI, the region of interest could be roughly segmented using a cursor. This was not hard to do because the lower bound of the soft palate in this slice direction was the oral cavity, which has low intensity, and the upper bound was the nasal cavity, which is also distinctive. In Livewire the user can guide the segmentation in real time, correcting any deviation from the region of interest easily. From the segmented area in this first slice, the median in each of the x, y directions are chosen to be the next slices to apply to Livewire. The similar approach is used to segment the soft palate from the coronal and axial slices. From the three segmented areas, a 3D rough active contour is constructed, where the volume inside the contour is given a value of 1 and a 0 elsewhere. Using this volume image as the active contour, a more satisfactory segmentation volume was obtained. Another segmentation with this approach was performed by using the a Gaussian Magnitude map produced by the GradientMagnitudeFilter method in matitk as the active contour. Results from using this gradient map produced a segmented region that is a lot smaller than the real region of interest. Livewire compliments the Geodesic Active Contour Segmentation algorithm well because the active contour does not have to be inside the region of interest; whereas the Confidence Connected Segmentation cannot be used in this case because the seed region needs to be within the region of interest. Livewire can be used to trace the edges around the region of interest, but does not extract a good seed region within the region of interest because the edges produces always surround the outer perimeter of the region and is not contained within it.. 5. FUTURE DIRECTIONSThe Livewire-Matitk combination method is only temporal because it requires too much human interaction. For batch processing of many data sets, this is not ideal. Using mesh data reconstructed from CT scans as boundaries has been suggested by Dr. Sidney Fels is a better approach. The mesh data could replace the Livewire method to produce a better active contour for the segmentation. This method is applicable because the Geodesic Segmentation method does not require the contour to be inside the region to be segmented, as do many segmentation algorithm. The mesh is the reconstructed representation of the rigid structure in the vicinity of the airway, including the hard palate, which serves as a boundary to the soft palate. Data points were simplified and output into Matlab format for use in the segmentation process. However, due to time constraints, they were not used. There are several complications to applying these boundary points that have not been resolved. First, the data points must be registered to the image of interest. Global scaling transformation can be applied to fit the mesh surface around the proximity of the soft palate region. However, only affine transformation at specified points can be used because the meshes represent bone structure so movement should be limited to joints. A good fit between the structures requires more work, but is not unachievable given more time. 6.RESULT IMAGE SAMPLESFigure 2: Registered image. a) Figure 3: Comparison of the image before and after smoothing. 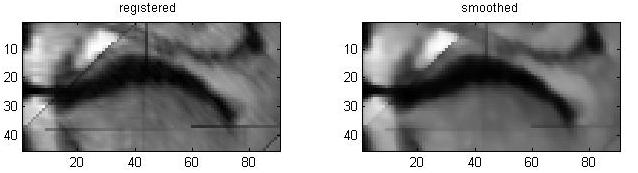 Figure 4: Segmentation using Connected Confidence (region growing) method. A point located near the centre of the soft palate volume was manually chosen as the seed region. a)Feature Image (smoothed) b)Sample slices of the output image c)3D contour slice representation of segmented region. The airway was segmented instead of the palate. 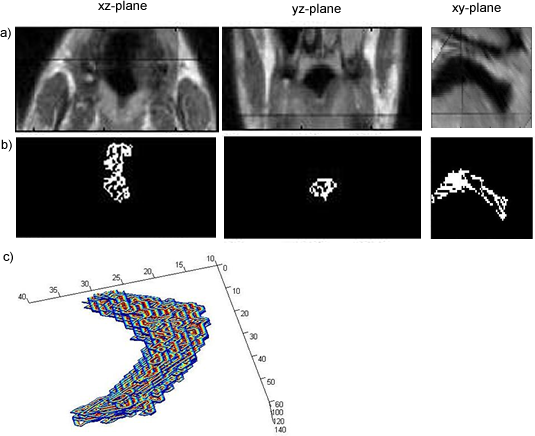 Figures 5,6,7: Segmentation using Geodesic Active Contours a)Feature Image (Preprocessed) b)Sample slices of the contour image used c)Sample slices of the output image Figures 5 and 6 : using Livewire output as contours Figure 5: smoothed by an Anisotropic Diffusion Filter Figure 6: gradient map calculated using the Gradient Magnitude Image Filter Figure 5: 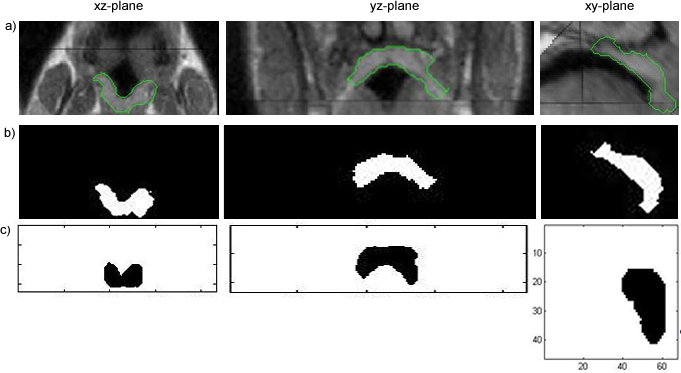 Figure 6: 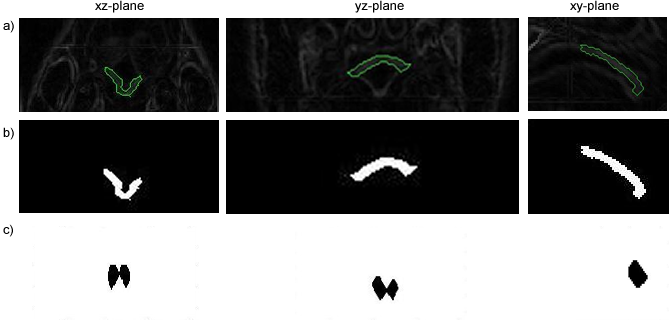 Figure 7:Using a Gradient Anisotropic Diffusion filtered output as gradient map 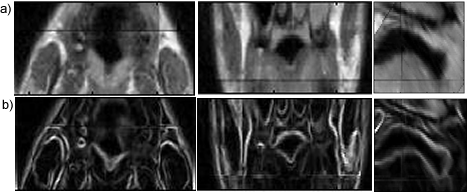 c) Segmentation was unsuccessful. No results obtained. Figure 8:3D contour slice representation of 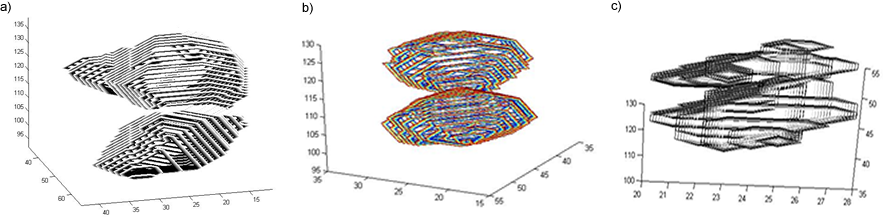 ACKNOWLEDGEMENTSTools used in this project such as Livewire, Matitk, and view3d were downloaded with premission from http://www.cs.sfu.ca/~hamarneh/software REFERENCES[1] L. Ib´a˜nez, W. Schroeder, L. Ng, J. Cates, and the Insight Software Consortium. The ITK Software Guide. Second Edition.Updated for ITK version 2.4. http://www.itk.org [2] M. Leventon,W. Grimson, and O. Faugeras. “Statistical shape influence in geodesic active contours”, in Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR),volume 1, pages 316–323, 2000. [3] G. Hamarneh, J.Yang, C. McIntosh, and M. Langille, “3D live-wire-based semi-automatic segmentation of medical images”, in Medical Imaging: Image Processing, Proc SPIE 5747, pages 1597-1603, 2005. [4] M. Kass, A. Witkin, and D. Terzopoulos, “Snakes: Active contour models,” International Journal of Computer Vision 1(4), pages 321–331, 1987. [5] A. X. Falcao and J. K. Udupa, “Segmentation of 3d objects using live wire,” in Medical Imaging: Image Processing, Proc. SPIE 3034, pages 228–235, 1997. [6] V.Chu,G. Hamarneh, “MATITK:EXTENDING MATLAB WITH ITK”. Taken from http://www.cs.sfu.ca/~hamarneh/software/matitk/ on November 23, 2007. |
|
| View Edit Attributes History Attach Print Search Page last modified on January 01, 2008, at 04:31 PM | ||
 b)
b)  c)
c)