|
|
||
| Menu | OPAL / ImagingAmpModeling | |
|
OPAL Home About People International Collaborators
Opportunities
Events
Related Links
|
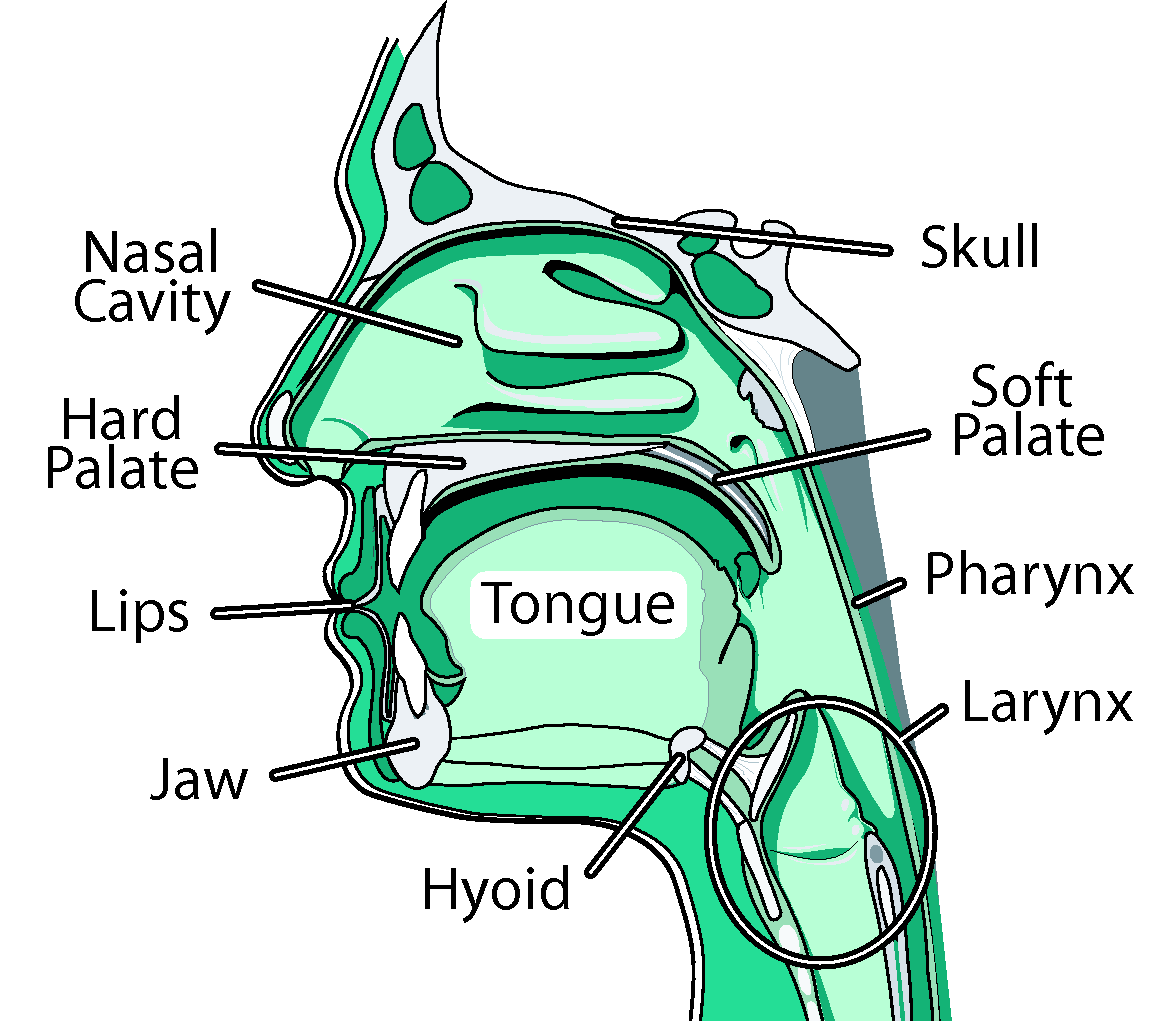
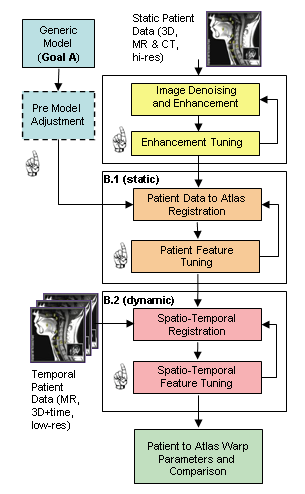
We are creating a comprehensive, adaptable, physically-based three-dimensional (3D) computer model of the human oral, pharyngeal and laryngeal (OPAL) complex (as shown in the figure), directed toward physiological research and clinical applications. We believe that physical simulations will become increasingly important for medical technology, permitting researchers to estimate quantities such as neuromotor activation levels or internal forces that are difficult to measure in vivo, and allowing clinicians to gauge the outcome of surgical procedures or formulate new treatments altogether. We believe modeling would improve our understanding of the biomechanics of airway collapse and could help in planning or developing treatments. For stroke-induced dysphagia, modeling would help us better understand how the neural-motor control of swallowing is affected by stroke, and could suggest therapies involving the stimulation of alternate neural pathways. Cancer-related surgical deficits arise when the removal of tissue for cancer treatment restricts the ability of patients to chew and swallow; here, simulation could permit better surgical planning and facilitate the design of prostheses. Other uses for our model are anticipated: as a training device, it will offer an immersive virtual environment that permits students to observe both static structures and dynamic behaviors (such as what happens when a particular muscle group is activated). Our efforts here have two main components: modeling and imaging. In our modeling efforts, we are developing a generic reference model of the OPAL complex anatomy using a mixture of medical image data and semi-automated segmentation methods. These models will be available to the research and medical communities through the ArtiSynth open-source software platform, providing a tool that can be extended or modified to suit different applications. Much as we have created a dynamic physical model of the jaw-tongue-hyoid model, we will continue to add and refine different components creating a library of upper airway anatomy. In our imaging efforts, we consider how to register reference models to fit specific patients. We are actively looking for graduate students and undergraduates who would like to participate in the activities below. Please contact us if you are interested in getting involved. Activities in the imaging and modeling area include: Reference Model ConstructionThe objective of this activity is to create a standard reference model of the OPAL complex, using imaging data from one or more prototypical subjects, that can be used for generic study or registered (or “morphed”) into a patient-specific model. Key sub-projects include: Structure IdentificationThis activity involves identifying the key model structures and their interconnections, driven by the clinical applications of this project. We are looking primarily at the jaw, skull, tongue, hard and soft palate, pharynx, lips, nasal cavity, larynx, and connecting tissue and muscles. Tools for improving segmentation workflow discusses some of the existing tools available for this activity. Geometry Extraction and Image Atlas CreationWe extract the geometry for most of the model’s components ourselves, using CT and MRI data acquired for prototypical subjects using facilities at UBC's MRI Research Centre http://www.mriresearch.ubc.ca/ and with Dentistry's Cone-CT scanner. This data is enhanced using appropriate methods such as field MRI intensity inhomogeneity correction or edge preserving non-linear diffusion, and then segmented using our recently developed, highly automated, 3D Livewire segmentation technique. Surface mesh geometry is extracted for hard and soft tissue components, in conjunction with other meshing tools (Amira, Rhino). Ultimately, labeled data sets are combined to form an image atlas, providing the basis for the patient-specific modeling. Some initial investigation of soft palate segmentation using different techniques such as: LiveWire, Region Growing and level set methods are discussed here. Dynamic modelingComponent geometries will serve as the basis for creating dynamic components to simulate actual physical behavior. We use a combination of rigid bodies (jaw, skull, hyoid, laryngeal components), deformable bodies (tongue, lips and soft palate), and deformable shells (pharynx). Muscle activity may be simulated using point-to-point forces emulating Hill or other activation models; these forces act between special marker points or between nodes sets within the deformable models. Patient-Specific Model RegistrationThis activity focuses on matching patient-specific medical imaging data to the reference models developed in the activities above, usually with the aim of deforming, or registering, it to form a patient-specific model. Typically, this process is complex and tedious, so this activity looks at ways to make it easier using a three-stage computational image analysis pipeline with stages of automatic processing and human intervention as shown in the figure. The sub-activities include: Static Registration of Patient-Specific ModelsThe objective here is to first create a patient-specific static model using the reference model and patient-specific imaging data. MRI and CT data similar to that used in reference model construction above are collected and enhanced as shown in the pipeline. A correspondence is automatically computed between this data and the reference model image atlas, yielding a deformation field which transforms all the geometric features of the reference model to the patient. Complete registration requires two steps: one for hard tissue (e.g., bone) and one for soft tissue, since each uses different transformation parameters. Temporal Registration of Image SequencesLow resolution (2D, 2.5D and 3D) temporal MR data of subjects chewing, swallowing, sleeping and speaking captures the high speed dynamics for investigating and clinical applications such as OSA and Dysphagia. To efficiently register this data, we begin with a static model and a deformation field which can be used to create a patient-specific image atlas. This is then down-sampled and matched to the closest-fitting low resolution image, with adjacent images then matched in succession, producing a temporally varying deformation field which captures the dynamics of moving structures. User-centred Design for Streamlining Patient RegistrationThe patient-specific modeling process described above involves complex user interactions employing many different tools. Our objective with this activity is to make this easier for a user with minimal technical training, which is necessary for deployment in clinical settings. |
|
| View Edit Attributes History Attach Print Search Page last modified on March 04, 2014, at 10:37 AM | ||