|
|
||
| Menu | OPAL / TriciaTPSWarping | |
|
OPAL Home About People International Collaborators
Opportunities
Events
Related Links
|
Thin Plate Spline Warping to Create a Patient-Specific ModelTricia Pang, Jul. 2008 Motivation Previous attempts of segmenting the tongue for patient-specific models, such as through using 3D Livewire, were found to involve too much manual work and/or produced a noisy result that was insufficient for import into the Artisynth environment. This article proposes a method for landmark-based spline deformation of a 3D geometric prior model to match a soft-tissue MRI volume of a specific patient, producing a patient-specific polygonal mesh. Creating a model of the tongue was the primary focus as it is one of the most important structures in the OPAL project and Artisynth tongue-jaw model, and it has a moderate-to-difficult shape. Algorithm The thin-plate spline (TPS) deformation is an algorithm first introduced by Duchon in 1976, and later redefined by Bookstein for interpolating surfaces over a set of landmarks. The landmarks, or scattered selected data points, define two sets of corresponding or homologous points that influence the degree and magnitude of deformation. Bookstein defines the spline deformation as a decomposition into a linear part and the superposition of “principal warps” [1]. The linear deformation is the application of an affine transformation: translation, rotation and shearing. The principal warp is the application of an affine-free local deformation of smaller geometrical scale based on radial basis functions. There are several spline-based deformations, but the TPS method was selected as it is known from registration studies in medicine [1] [2] [3] [4]. Bookstein's TPS method was defined for 2D and implemented for use in Matlab by Dollar and Rabaud in their Image and Video Toolbox for Matlab [5]. This project involved extending Dollar and Rabaud's implementation to 3D, with an algorithm based on the 3D regularized implementation described by Muller, Mang and Buzug [4]. A TPS deformation field is generated when given a set of corresponding landmarks between the reference model and the MRI data set. The deformation field can then be applied to all the points in the reference model, interpolating them to fit the specified points in the MRI data set. Design The "Corresponding Point Selection" (CPS) graphical user interface was created in Matlab to allow a user to manually select landmark points in a set of MRI images and their corresponding points on an 3D geometric reference model. 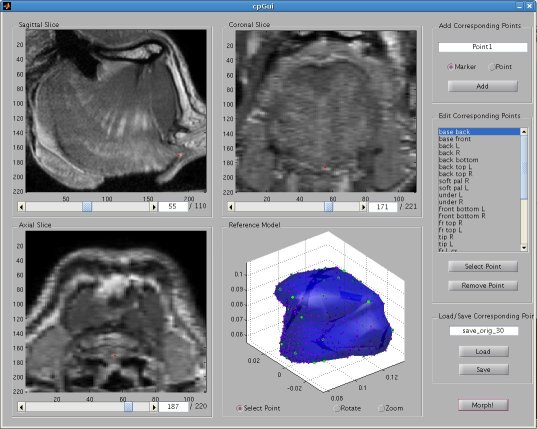 Graphical user interface for selecting corresponding points between an MRI data set and a reference model mesh In selecting a point on the MRI set, the three different orthogonal slice views are shown (sagittal, axial and coronal). When the user clicks on a point on any one of the views, the corresponding two slices that intersect that point is immediately displayed on the other two views to help the user gauge the location of his click point. The slices can also be navigated using a scroll bar or manually entering a slice number. The reference model used was an ICP tongue model exported from Artisynth. By using this tongue model, the patient-specific model resulting from warping can be easily imported back into the environment. In Artisynth, a Jython script was implemented to export the model's vertices, surface faces that connect vertices, muscles situated between vertices, and the activation of each muscle. Prior to exporting the model, the g-force and muscle excitations were modified to simulate a person lying down, to try and replicate the subject's position in the MRI scanner. When imported into the CPS tool, the reference model is displayed as a 3D mesh, and buttons allow the user to rotate, and zoom in and out on the model. When the user clicks on a point on the model, the vertex closest to that point is selected. When a point is selected on the MRI set and a point is selected on the reference model, the point can be given a description and added to a list of corresponding points. Points added to this list can be selected for reviewing or deletion. The list of user-selected corresponding points can also be saved to disk or loaded from disk. After the user has completed selecting a set of corresponding points, the result can be generated. First, the MRI is normalized to the same scalar voxel dimension as the reference model. Currently the MRI is translated so that its centroid is at the centroid of the reference model to ensure the MRI is in approximately the correct location on import into Artisynth. (In the future, landmarks should be exported from Artisynth as well to pre-register the location where the result model should reside.) The thin-plate spline implementation described above in the Algorithm section is then applied and the result is exported. Result Using 30 scattered corresponding points, the average discrepancy between the 30 selected destination points and the location of the 30 points after warping was negligible, at 2.6 x 10^-17 mm. The movement of the points on the reference model to create the warped model is shown below. 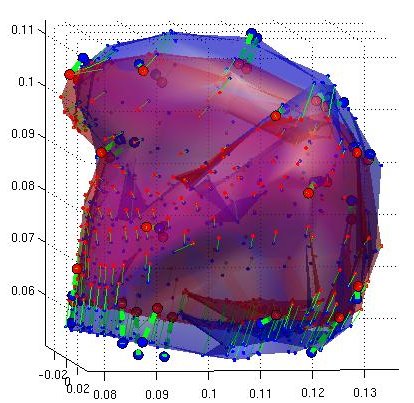 Overlay of reference model (red mesh) on the warp result (blue mesh), showing warp source points (red circles) and destination points (blue circles), and the resulting warp field (green lines) from the movement of each point The result from warping was also compared to the result from segmentation using 3D Livewire. The average discrepancy between each resulting point from warping and its closest point found on the 3D Livewire result was 5.6 x 10^-7 mm. In comparison, the average movement of the 30 selected points was 1.0 x 10^-5 mm -- this gives the warping result an error of 0.056%. When overlaying the resulting 3D mesh from warping over the Livewire result, there are several noticeable differences. As noted in Muller, Mang and Buzug's work [4], TPS warping is not able to transform a model beyond its original topology, so a MRI soft-tissue database is needed containing different reference models for distinct subject differences such as in weight and physiognomy. Moreover, the tongue template must have the same structure from the tongue in the MRI data, which will involve decisions on whether to include certain components such as glands. The template must also be based on data from a patient in the same posture (lying on his back with tongue pressed against the hard palate). The noisy result from 3D Livewire is also evident, and likely increases the error result. A manually segmented model would be better in representing the absolute truth when assessing the warp result. 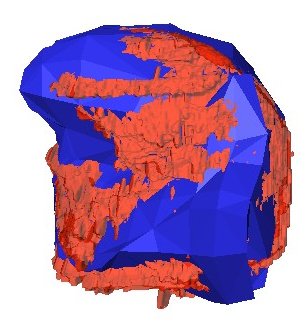 Overlay of warp result (blue mesh) on the 3D Livewire result (red volume) Suggestions for Future Work
References [1] Bookstein, F. L., "Principal Warps: Thin-Plate Splines and the Decomposition of Deformations," IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 11, no. 6, pp. 567-585, Jun. 1989. [2] Carr, J. C., Fright, W. R., and Beatson, R. K., "Surface Interpolation with Radial Basis Functions for Medical Imaging," IEEE Transactions on Medical Imaging, vol. 16, no. 1, pp. 96-107, Feb. 1997. [3] Fang, S., Raghavan, R., and Richtsmeier J. T., "Volume Morphing Methods for Landmark-Based 3D Image Deformation," Proc. SPIE 1996, vol. 2710, pp. 404-415, Apr. 1996. [4] Muller, J., Mang, A., and Buzug, T. M. "A Template-Deformation Method for Facial Reproduction," Proc. ISPA 2005, pp. 359-364, Sep. 2005. [5] Dollar, P., and Rabaud, V., "Piotr's Image & Video Toolbox for Matlab," http://vision.ucsd.edu/~pdollar/toolbox/doc/, ver. 2.11, Jul. 2008. |
|
| View Edit Attributes History Attach Print Search Page last modified on July 31, 2008, at 04:39 PM | ||